Blossom Algorithm
The blossom algorithm, sometimes called the Edmonds’ matching algorithm, can be used on any graph to construct a maximum matching. The blossom algorithm improves upon the Hungarian algorithm by shrinking cycles in the graph to reveal augmenting paths. Additionally, the Hungarian algorithm only works on weighted bipartite graphs but the blossom algorithm will work on any graph. The blossom algorithm is a polynomial time maximum graph matching algorithm. This algorithm has many applications, for example, assignment problems such as the Hall's marriage theorem, and path problems such as the Hamiltonian path problem.
Contents
Motivating The Algorithm and Intuition
The Hungarian algorithm is a very efficient graph matching algorithm, but it only works on weighted bipartite graphs as it cannot deal with odd length cycles. The issue lies in finding augmenting paths, since if there is an odd cycle, the algorithm will calculate the wrong augmenting path because it will miss some of the edges. Additionally, a cycle may make the algorithm loop forever, reducing, in the case of minimum-weight optimization, the weight infinitely many times, inhibiting proper termination. The Blossom algorithm aims to fill this gap by solving more generalized graph matching problems, and can be used when the graph is not guaranteed to be bipartite.
This algorithm improves the Hungarian algorithm by having a way to deal with odd-length cycles. It does this by finding a way to trick the algorithm into thinking it is operating on a simple bipartite graph, collapsing a cycle into a single vertex, so it can run the Hungarian algorithm on the matching.
The algorithm is named as such since a cycle looks much like a blossom, alluding to a flower. These cycles can be shrunk and the search is then restarted recursively. If the algorithm finds an augmenting path in a shrunken graph, it can be expanded up through the blossoms to yield an augmenting path in the original graph. This alternating path can be used to augment the matching by one edge. If the recursive process ever runs into a state where there is no augmenting path, then the algorithm can return that there is no augmenting path in the original graph.[2]
The Blossom Algorithm
The blossom algorithm takes a general graph \(G = (V, E)\) and finds a maximum matching \(M\). The algorithm starts with an empty matching and then iteratively improves it by adding edges, one at a time, to build augmenting paths in the matching \(M\). Adding to an augmenting path can grow a matching since every other edge in an augmenting path is an edge in the matching; as more edges are added to the augmenting path, more matching edges are discovered. The blossom algorithm has three possible results after each iteration. Either the algorithm finds no augmenting paths, in which case it has found a maximum matching; an augmenting path can be found in order to improve the outcome; or a blossom can be found in order to manipulate it and discover a new augmenting path.
Augmenting Paths
An augmenting path is an odd length path that has unmatched nodes for endpoints. The edges in an augmenting path alternate: one segment will be in the matching, the next segment is not in the matching, the next segment is in the matching, and so on.
The algorithm works by finding augmenting paths so it can increase the size of the matching by one each iteration. To do this, it starts by finding unmatched vertices (vertices that are not connected to an edge that is in the matching). Then, it builds up an alternating path from these nodes. This means that the path begins at a node not in the matching — to be an augmenting path, it must start and end with unmatched nodes.
The algorithm continues to build on the augmenting path until it can’t anymore. When it stops building an augmenting path, it is either the case that the graph has run out of edges that are not already in the augmenting path, in which case, there is no maximum matching, or the algorithm gets to a point where an augmenting path cannot be made, at which point the algorithm returns a maximum matching.
Blossoms
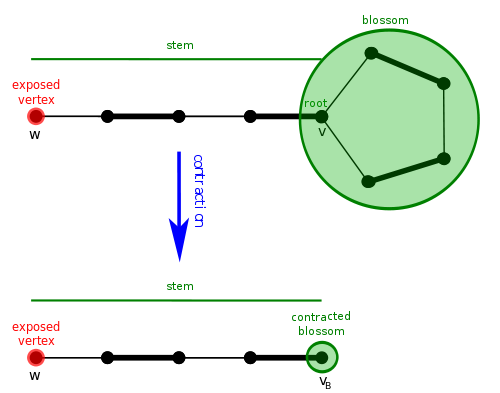
The idea behind blossoms is that an odd-length cycle in the graph can be contracted to a single vertex so that the search can continue iteratively in the now contracted graph [3]. Here, these odd-length cycles are the blossoms. The algorithm can just go on through the graph and treat the tricky cycle as if it were only a single node — fooling the algorithm into thinking there is an even number of nodes in the graph (it has explored so far) so that it can run the Hungarian algorithm on it.
A blossom is a cycle in \(G\) consisting of \(2k + 1\) edges of which exactly \(k\) belong to \(M\), and where one of the vertices \(v\) in the cycle (the base) is such that there exists an alternating path of an even length, called a stem, from \(v\) to an exposed vertex \(w\)[3]. The advantage is given due to the ease at which the stem part can be toggled between edges in the blossom.
The goal of the algorithm is to shrinks a blossom into a single node in order to reveal augmenting paths. It works by running the Hungarian algorithm until it runs into a blossom, which it then shrinks down into a single vertex. Then, it begins the Hungarian algorithm again. If another blossom is found, it shrinks the blossom and starts the Hungarian algorithm, and so on. [4]
 Notice the toggling in edges to make another matched edge the stem of the blossom[5].
Notice the toggling in edges to make another matched edge the stem of the blossom[5].
Pseudocode
Formally, the algorithm is performed by maintaining a forest of scanned edges.
\(M = 0,\ H = G,\ N = M\)
EVEN = { v | v is free in M}, ODD = 0, \(E_f = 0\), \(F = (V_F, E_F) \).
Maintain \(V_F = EVEN \cup ODD \).
Pick an edge \(u, v\) such that \(u\ \epsilon\ EVEN\) and \(v\ \notin\ ODD\)
a. If none exist, output \(M\).
If \(v\ \notin\ V_F\) then:
a. Put \(v\) in ODD, \(M(v)\) in EVEN, and \( (u, v) \) and \( (v, M(v)) \) in \(E_f\), where \(M(v)\) is the vertex matched to \(v\).
Else, if \(v\ \epsilon\ EVEN\) and \(u, v\) are connected in \(F\), then:
a. A blossom \(B\) has been found.
b. Shrink \(B\) to \(b\), and set \(H\) to \(H \setminus B\), \(N\) to \(N \setminus B\).
c. In \(H\), put \(b\) in EVEN.
d. Go to step 4.
Else, if \(v\ \epsilon\ EVEN\) and \(u, v\) are not connected in F, then:
a. There is an augmenting path \(\nu\) in \(H\) with respect to \(N\).
b. Retrieve an augmenting path \(\rho\) in \(G\) with respect to \(M\).
c. Augment \(M\).
d. Go to step 2.
End algorithm when no more conditionals are satisfied.
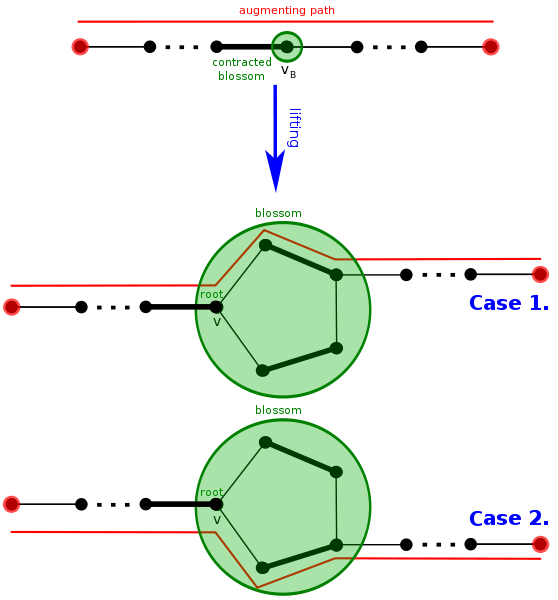
In the pseudocode above, a blossom can be easily spotted if two vertices connected by an edge are EVEN vertices, implying that the cycle is odd. Similarly, an augmenting path \(\rho\) can be retrieved by unshrinking the blossom and matchings shrunk along the way. If an augmenting path in \(H\), \(\nu\), is obtained from \(G\) by shrinking \(B\) to \(b\), if \(\nu\) does not visit \(b\), then \(\nu\) is also an \(M\) augmenting path. Otherwise, either \(\nu\) ends in b, in which case the blossom \(B\) included the root of the tree so \(\nu\) ends at some vertex \(B\); or \(\nu\) visits \(b\), in which case \(\nu\) reaches vertex \(x\) on \(B\), and the blossom can be altered so that the matched edge can connect the graph with the root, as seen in Image 6 above [7].
Complexity
The blossom algorithm has three outcomes contributing to the runtime complexity: it may find a maximum matching, an augmenting path, or a blossom. In the worst case, this algorithm has a total runtime of \(O(|E||V|^2)\), where each iteration takes \(O(|E|)\) as each edge is scanned in hopes of asserting a perfect matching. If an augmenting path is found, the matching can be reconfigured in \(O(|V|)\) time, taking \(O(|E||V|)\) time. Lastly, if a blossom is found, the smaller graph has to be recursed, which can occur repeatedly for a maximum of \(O(|V|)\) recursions, at which point either an augmenting path is found, or the algorithm terminates. Since each time a blossom is recursed, there is a new set of augmenting paths, the algorithm can take a maximum runtime of\(O(|E||V|^2)\) [8].
This algorithm was later improved to have a runtime of \(O(|V^3|)\), and subsequently further reduced to \(O(|E|\sqrt{|V|})\). For further details on the fast implementation refer to Micali and V. Vazirani’s paper, A Proof of the MV Matching Algorithm.
See Also
References
- , A. Edmonds blossom.svg. Retrieved June 19, 2016, from https://en.wikipedia.org/wiki/File:Edmonds_blossom.svg
- Wagon, S. Blossom Algorithm. Retrieved June 26, 2016, from http://mathworld.wolfram.com/BlossomAlgorithm.html
- , . Blossom Algorithm. Retrieved June 19, 2016, from https://en.wikipedia.org/wiki/Blossom_algorithm
- Kusner, M. Edmonds’ Blossom Algorithm. Retrieved June 19, 2016, from http://matthewkusner.com/MatthewKusner_BlossomAlgorithmReport.pdf
- , A. Edmonds lifting path.svg. Retrieved June 25, 2016, from https://en.wikipedia.org/wiki/File:Edmonds_lifting_path.svg
- , A. Edmonds lifting path.svg. Retrieved June 25, 2016, from https://en.wikipedia.org/wiki/File:Edmonds_lifting_end_point.svg
- Mahajan, M. Matchings in Graphs. Retrieved from http://www.imsc.res.in/~meena/matching/edmonds.pdf
- Gupta, A. Lecture #8: Edmond’s Blossom Algorithm. Retrieved from http://www.cs.cmu.edu/~anupamg/advanced/lectures/lecture08.pdf