Strings
Strings are ordered sets of characters, which are used to represent all sorts of non-numerical data such as works of literature, genetic sequences, encrypted messages of great importance to the future of mankind, or the names of people on your softball team. By digitizing textual sorts of data, many incredible tasks can be performed from contextually searching your computer's filesystem, to tracing the evolutionary lineage of organisms, to the instant decryption of ancient ciphers. Order is important and distinguishes strings from sets of characters.
 wikipedia
wikipedia
Basic String Features
Strings are a built-in data type in most modern languages and come equipped with a number of useful operations. Common tasks with string data require us to dissect and recombine them, and find patterns within them. Pattern finding in strings can get sophisticated quickly, and more powerful string-matching techniques are relegated to the regular expressions article. Below, we'll explore some of the fundamental manipulations that can be performed with strings.
Character extraction
Each character in a string is given a nice address known as an index that can be used to retrieve it. The usual convention is for the first character (starting from the left) in a string to have the index 0, the next has index 1, and so on. In many languages, strings can also be indexed from the right-hand side, using negative numbers to count. The last position (again counting from the left) is assigned -1, the second rightmost character is indexed by -2, and so on until the beginning of the string is reached. Obviously, the maximum index in a string is its length minus one. Similarly, its lowest index is minus the length, which is the index of the first element.
For instance, in the string
"Merriweather"theMhas index0from the left and-12from the right.An example is worth many words, and let's see how character extraction works in Python.
> s = 'abcdef' > print s[4] # print the character residing at index 4. 'e'
Substring extraction
The index can be used to extract a substring out of the string if we know the starting and ending index of our sub-string.
> s = 'abcdef' > print s[1:4] 'bcd'It is important to note that the resulting sub-string ends one index before the upper index.
What number will the following Python code print?
1 2 3 4 5 6 7 8 9 | |
Character extraction
It is possible to simultaneously use the positive and negative indexing schemes.
> s = 'abcdef' > print s[-1] # print the last character. 'f' > print s[2:-1] # print from index 2 up to (but not including) the last index. 'cde'Again, it's important to note that the substring ends on the character before the end of the range (in Python, slicing a list/string is [inclusive:exclusive], so
:-1will actually include the character at index-2but not-1).The convenient thing about negative indexing is that one need not know the length of string in order to obtain characters from the end.
Once we extract substrings from a string, it is often necessary to recombine pieces into yet another string. Most programming languages include an operator for joining two strings.
String concatenation
In Python, we can concatenate strings with
+operator.
>str1 = 'Good' >str2 = 'morning' >str3 = 'Vietnam!' >print str1 + ' ' + str2 + ' ' + str3 Good Morning Vietnam!
Substrings
Continuing our idea of using ordered group of characters which we have named "strings," we will be learning more about them to be able to do powerful things with them and finally solve real-life computation problems.
Strings are simple in definition, but that doesn't mean they have limited applicability. They are basic building blocks which can be used to build more sophisticated data types to solve problems. Familiarity with string manipulation is important because many applications require one to play with strings and reshape them into a form that's useful for computation.
Below are some useful elementary string functionalities:
Python Substring Method
In addition to being able to grab a substring using the
[start_inclusive:end_exclusive]notation, a third parameter can be provided to add a "step" size which will periodically sample the region between the start and end indices. By default, the starting and ending indices of the substring method are0andlen(str); thus if we ask for[:], we obtain the whole string.For example:
> str = 'abcdefghij' ######## 0123456789 > str[1:8:2] # A step size of 2 prints every second character in the substring. # Only characters appearing at indices 1, 3, 5, and 7 will be printed. 'bdfh' > str[::-2] # A step size of -2 prints every 2nd character in the substring, counting # backward from the end of the string. Only characters appearing at indices # -1, -3 -5, -7, -9 will be printed. 'jhfdb'Pretty simple!

Pantha du Prince sends you a secret message hidden within a string of nonsense. You know that the real message is periodic within the garbage and that he's partial to the number three.
Find Pantha's hidden message to you by taking periodic substrings.
panthas_message = "Tkxhwbiscscu vfbmwlgzirlsnysqr"
Splitting
Because we often receive strings of variable length, indices will not always be useful. A common form of data storage involves separating items by a universal separator such as a comma, spaces, tabs, underscores, etc. In any separator scheme, users must agree not to use the separating string as an informative part of their file, i.e. the separator is only ever used to demarcate boundaries of items, but not items as such.
As this is common, a method to break strings up at the site of such characters is provided in most languages. Given an input string and the identity of the separator, the split method will return the component strings.
Python string split() method
Suppose you have a file that contains strings of the form below, representing various gene products:
line =" lacZ_1013aa_betagalactosidase_escherichiacoli"where the first string is the name of the gene, the second is its length in amino acid residues, the third its enzyme product, and the fourth its organism of origin.
Write a function that breaks up the input string and prints the enzyme product.
First, we can use the
splitmethod to breaklineup into the four fields, which yields an array of strings. Then we simply print the third element.
def get_enzyme(ln): splitted = ln.split('_') enzyme_product = splitted[2] print enzyme_productThus
>line = lacZ_1013aa_betagalactosidase_escherichiacoli >get_enzyme(line) "betagalactosidase"
Translation
Another common task with strings is to make character substitutions, where we replace characters x1, x2, ..., xn with an alternative alphabet y1, y2, ..., yn. For example, suppose we have a region of the genome (DNA) corresponding to gene X, whose product is protein X, and we want to find the mRNA used by the ribosome to translate the information into protein.
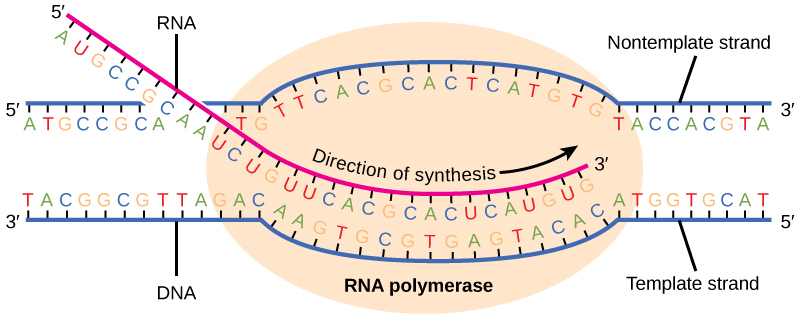
RNA polymerase reads through the negative strand and replaces all cytosines with guanine, and vice versa. All thymines are replaced with adenosine. And, as the base uracil is used in place of thymine in all RNA sequences, adenosine is replaced with uracil. Thus we have
1 2 3 4 | |
How can we transcribe the DNA sequence into mRNA using string translation?
RNA transcription
We can use method
maketransfrom the Python string module to make a Rosetta stone for this translation task:
>from string import maketrans >rosetta = maketrans('ATGC', 'UACG')With the translation table in hand, we can use it to transcribe our DNA sequence into an RNA. Due to the 5' -> 3' polarity of nucleic acid sequences, this will be the reverse of our goal, the mRNA sequence:
>DNA = "GCGTGAGTACACATGGTGCAT" >rev_mRNA = DNA.translate(rosetta)Finally, to obtain the mRNA sequence, we reverse our result:
>mRNA = rev_mRNA[::-1] 'AUGCACCAUGUGUACUCACGC'In the last step, we've used the substring functionality to obtain the whole string, with the index running in reverse.